The number and complexity of impacting questions in clinical and healthcare envinroment gives rise to an unprecedented call to arms in which engineering, analysis and physics are requested to take part along with medical expertise for impacting clinical practice and its diagnostic, predictive and therapeutic purposes. In fact, current clinical practice would benefit from intelligent support systems to properly exploit the consistent amount of data that are generated on a daily basis, particularly on those tasks that can be further improved and optimized – i.e., early diagnosis (in terms of accuracy and velocity), patients monitoring and personalization of the therapeutic pathways.
Specifically, biomedical signals are observations of physiological activities of organisms, ranging from gene and protein sequences, to neural and cardiac rhythms, to 2D and 3D body images (PET, CT, MRI). Once ingested, medical unstructured data need to be integrated and processed to build models for complete patient profiling, treatment planning and response prediction models.
Efforts in this direction are mainly devoted to investigating imaging analysis methods to address clinical research questions, including but not limited to statistical (i.e., radiomics), geometrical, structural, transform-based and multivariate image analysis as well as image embedding, image semantics and convolutional neural networks. Imaging analysis has evolved into a discipline and been shown to represent a fundamental source of information when performing diagnosis, modelling disease progression, and predicting disease free survival.
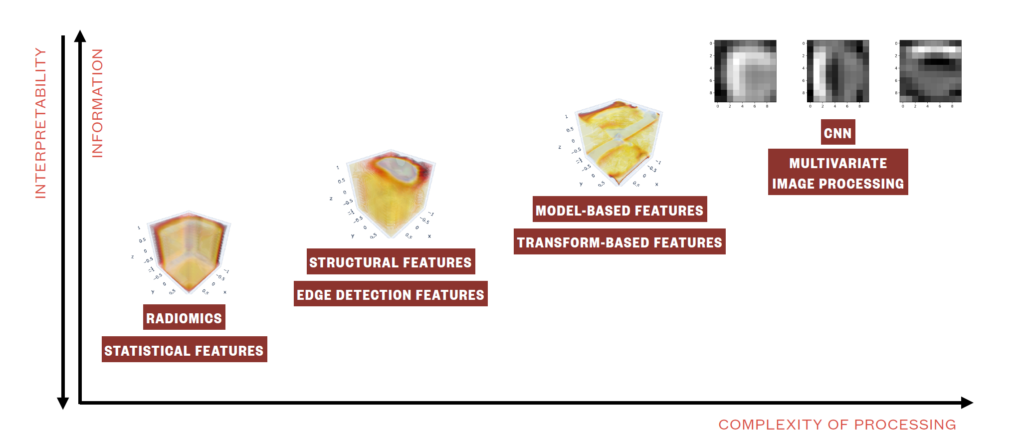
Our group tackle the methodological problems under different perspectives, both with supervised and unsupervised approaches, reaching insightful knowledge on the clinical and translational side. Accordingly, several novel methodologies have been and are being explored and developed, positioning on the cutting-edge of current literature.
Research projects/lines:
- Humanitas – Lymphoma Project

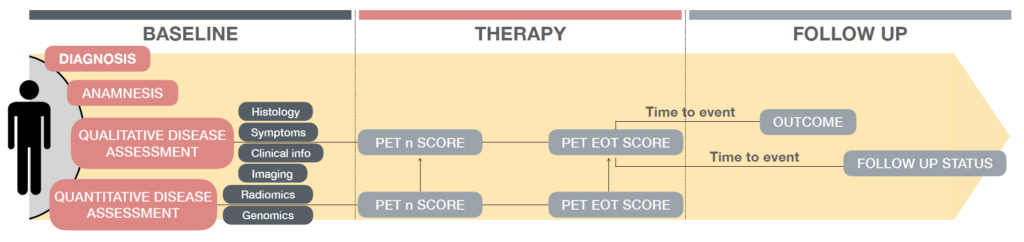
Goals: The project aims to elucidate the yet unknown Hodgkin Lymphoma underpinnings and to develop criteria for therapy response prediction through tumor imaging assessment and intra-tumor heterogeneity quantification.
Clinical partners: Dr. Arturo Chiti, Dr. Martina Sollini, Dr. Carmelo Carlo-Stella (Humanitas University), Dr. Margarita Kirienko (Istituto Nazionale dei Tumori)
- Humanitas – ViBE Project

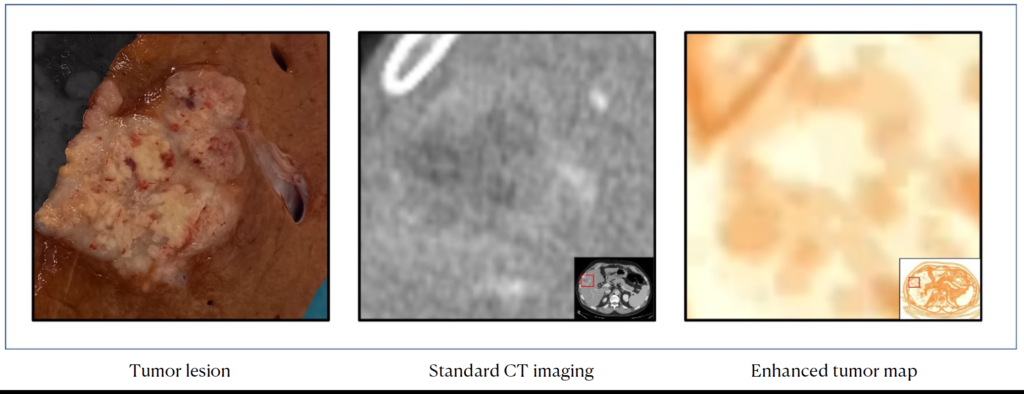
Goals: The project aims to build a liver virtual biopsy tool for cancer profiling. It consists of a processed and navigable 3D map of the tumor as to make imaging data easy to interpret for clinicians by visual inspection, disclose any intra-tumoral heterogeneity, that can be further quantitively characterized, and elucidate the association between heterogeneity and pathology and survival data, towards a full clinical translation or radiomics.
Clinical partners: Luca Viganò, Guido Costa, Francesco Fiz (Humanitas University)
- Pisa – Prostate Project

Goals: The project aims to provide a patient representation of multi-lesion disease, modelling information in an insightful, agnostic and easy to visualize way as to quantify tumor heterogeneity and predict tumor progression and treatment response in prostate cancer patients.
Clinical partners: Dr. Paola Anna Erba, Dr. Roberta Zanca (University of Pisa)
- Embedding and Cross-Subject Channel Selection Algorithm for Trial Classification

Goals: EEG Channel Selection is the process of identifying the most relevant electrodes to record EEG signals useful to discriminate specific brain activities. Most EEG systems are multi-channel in nature, but multiple channels might include noisy and redundant information and increase computational times of automated EEG decoding algorithms. To reduce the signal-to-noise ratio, improve accuracy and reduce computational time, one may combine channel selection with feature extraction and dimensionality reduction. However, as EEG signals present high inter-subject variability, we introduce a novel algorithm for subject-independent channel selection through representation learning of EEG recordings.
The algorithm exploits channel-specific 1D-CNNs as supervised feature extractors to maximize class separability and reduces a high dimensional multi-channel signal into a unique 1-Dimensional representation from which it selects the most relevant channels for classification. The algorithm can be transferred to new signals from new subjects and obtain novel highly informative trial vectors of controlled dimensionality to be fed to any kind of classifier.
- PROMETEO project

Goals: Anticipating diagnostic time, reducing infarction complications and optimizing the number of hospital admissions are three main goals of this project. Thanks to the partnerships of Azienda Regionale Emergenza Urgenza (AREU) and Abbott Vascular, ECG machinery with GSM transmission have been installed on all Basic Rescue Units of Milan urban area. PROMETEO project, planned and realized by 118 Dispatch Center of Milan since the end of 2008, made possible to send quickly the ECG from territory to 118 Dispatch Center itself, and then to the hospital where patient would have been admitted to, even when a Basic Rescue Unit (Unit managed by volunteers only, without physicians on board) is sent to the patient.
Clinical partners: AREU 118, Dr. Niccolò Grieco, Dr. Giovanni Sesana (Niguarda Ca’ Granda Hospital)
People involved:
- Prof. Francesca Ieva
- Dr. Lara Cavinato
- Dr. Michela Massi
- Dr. Alessandra Ragni